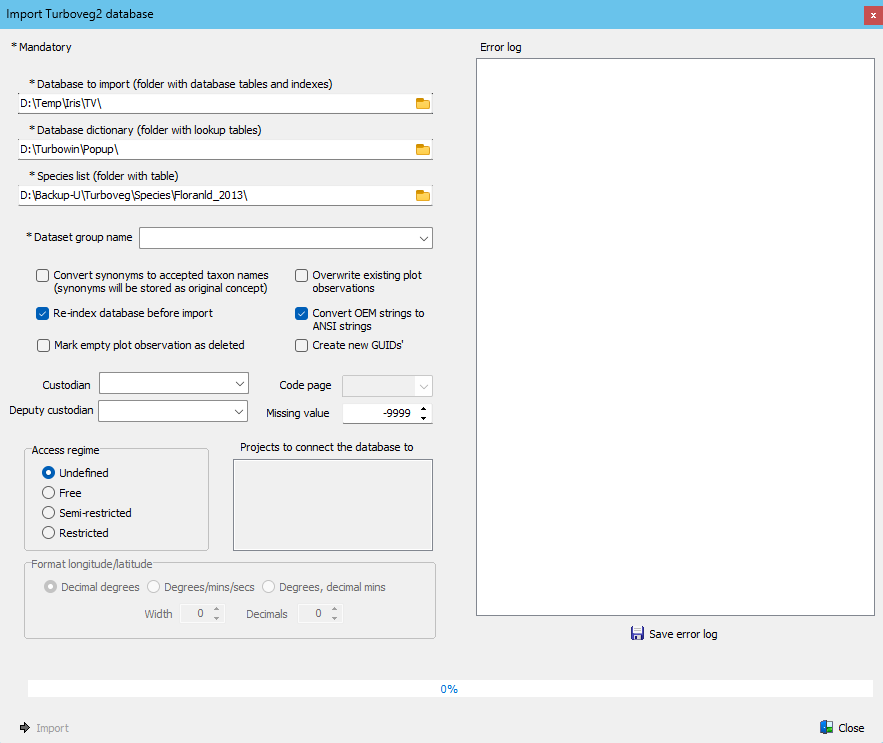
Single Turboveg2 databases can easily be imported in a Turboveg3 database. The import procedure is very strict when it comes to codes that are represented in look up tables (e.g. cover scales, authors, projects, species). Codes that are not present in a lookup table will be ignored, and an error message will pushed to an error message log. The logging information can be used to improve the Turboveg2 database which can then be re-imported in the Turboveg3 database.
•* Database to import: Full path of the Turboveg2 database to import.
•* Database dictionary: Full path to the corresponding Turboveg2 dictionary (set of lookup tables; 'popup lists' in Turboveg2).
•* Species list: Full path to the Turboveg2 species list connected to the database.
•* Dataset group name: Select the name of the dataset group in the Turboveg3 database to which the dataset needs to be assigned.
•Convert synonyms to accepted taxon names: This conversion can be useful for example when hunting for duplicated plots. Note that synonyms are stored as 'original species concept'.
•Re-index database before import: Check this option if you suspect that one of the Turboveg2 index files is corrupted.
•Convert OEM strings to ANSI strings: Uncheck this option if an Oem to Ansi conversion of strings is not required, otherwise check this option. This option needs trial and error in case special characters like 'š' do not appear properly on the screen.
•Overwrite existing plot observation: In case plot observations ID's need to be preserved and a certain dataset needs to be updated, this option should be checked. Otherwise the plot observation will not be imported as a unique index on the plot observation GUID's prevent double entries. With the option checked, Turboveg will first delete the old plot observation and then import the updated version.
•Mark empty plot observation as deleted: Mark plot observation as deleted automatically if they are empty (no species records available).
•Create new GUIDs': Only check this option if you are sure that not all plots in the Turboiveg2 database are unique. Warning. Before the actual import takes place the dataset with the same name as the Turboveg2 database will be deleted.
•Custodian: Select the custodian of the database to be imported. The list should be updated in the Turboveg3 database prior to the import. See Manage custodians.
•Deputy custodian: Select the deputy custodian of the database to import. The list should be updated in the Turboveg3 database prior to the import. See Manage custodians.
•Code page: Do not select a code page if the Turboveg databases are managed on your own computer.
•Missing value: Enter an number here, if a certain value is used to indicate missing values in the header data table of a Turboveg2 database. E.g. -9 or -9999.
•Access regime: All plots of a Turboveg2 database can be assigned to a selected access regime. The assignment is used to determine the policy for data exchange. The access regime is typically used for bigger database, like EVA, where many different data sources (from many different custodians) are brought together. This setting can be overruled by the value stored in the header data field 'EVA_ACCESS' in the Turboveg2 database, that allows the assignment of an access regime for each plot individually.
•Projects to connect the database to: Check one or multiple projects to which the dataset needs to be connected. This list of projects need to be updated in the Turboveg3 database prior to the import.
•Format longitude/latitude: Select 'DecimalDegrees' (default) if longitude and latitude are stored in decimal degrees in the Turboveg2 database. If 'Degrees/mins/secs' has been selected the Width and number of Decimals has te be set (check the values in the Turboveg v2 database). Note that the names of the fields that hold the longitude and latitude value should be LONGITUDE and LONGITUDE.
•Save error log: Save the error log to a file.
Dutch database
When importing Turboveg2 databases in a Dutch Turboveg3 database the following Turboveg2 fields are ignored:
SYNTAXON (Westhoff code, SBBCODE, ASSOCIA_02, ASSOCIA_03, ASSOCIA_04, ASSOCIA_05, INCOMPL_02, INCOMPL_03, INCOMPL_04, INCOMPL_05, WEIRDNE_02, WEIRDNE_03, WEIRDNE_04, WEIRDNE_05, NORM_LH_02, NORM_LH_03, NORM_LH_04, NORM_LH_05.
The field 'NEW_SYNTAX', that holds the Schaminée code, is renamed to SYNTAXON.